pygimli.physics.ert#
Electrical Resistivity Tomography (ERT) including IP.
Direct-Current (DC) Resistivity and Induced Polarisation (IP)
This package contains tools, modelling operators, and managers for Electrical Resistivity Tomography (ERT) & Induced polarization (IP)
Main entry classes:
DataContainer(ERT) - ERT/IP-specific data container
ERTModelling - Modelling operator
ERTManager - data inversion and modelling for real resistivity
ERTIPManager - extension to IP (either frequency or time domain)
TimelapseERT - processing and inversion of timelapse ERT data
CrossholeERT - timelapse ERT in crosshole environments
and functions:
simulate - synthetic (real or complex-valued) modelling
createData - generate data sets for synthetic modelling
load - load/import data from file supporting various formats
show/drawERTData - show ERT data as pseudosection or crossplot
generateDataPDF - generate a PDF file showing all data fields
createGeometricFactors - geometric factors analytical/numerical
estimateError - estimate data errors
reciprocalProcessing - fit error model through reciprocals
combineMultipleData - homogenize data containers
Overview#
Functions
|
Combine multiple data containers into data/err matrices. |
coverageDCtrans( (object)S, (object)dd, (object)mm) -> object : |
|
|
Utility one-liner to create a BERT datafile |
|
|
|
Create geometric factors for a given data scheme. |
|
Create default mesh for ERT inversion. |
|
Plot ERT data as pseudosection matrix (position over separation). |
|
Estimate relative error based on relative and absolute parts. |
|
Fit an error by statistical normal-reciprocal analysis. |
|
Generate data container from unique index. |
|
Generate a multi-page pdf showing all data properties. |
geometricFactors( (object)data [, (object)dim=3 [, (object)forceFlatEarth=False]]) -> object : |
|
|
Helper function to calculate configuration factors for a given DataContainerERT |
|
Shortcut to load ERT data. |
|
Return indices for reciprocal data. |
|
Reciprocal data analysis and error estimation. |
|
Remove duplicate rows from an ERT datacontainer. |
|
Plot ERT data as pseudosection matrix (position over separation). |
|
Plot ERT data as pseudosection matrix (position over separation). |
|
Plot ERT data as pseudosection matrix (position over separation). |
|
Simulate an ERT measurement. |
|
Generate unique index from sensor indices A/B/M/N for matching. |
Classes
|
Class for crosshole ERT data manipulation. |
|
Method manager for ERT including induced polarization (IP). |
|
ERT Manager. |
|
Forward operator for Electrical Resistivity Tomography. |
|
Reference implementation for 2.5D Electrical Resistivity Tomography. |
|
Class for crosshole ERT data manipulation. |
Functions#
- pygimli.physics.ert.combineMultipleData(DATA)[source]#
Combine multiple data containers into data/err matrices.
Generates a unified data container and a data matrices Non-existing data are filled with NaN values.
- Parameters:
DATA (list of DataContainerERT) – data contea
- Returns:
data (pg.DataContainerERT) – ERT data container with quadrupols from all input data
RHOA (np.array of size data.size() x len(DATA)) – apparent resistivity data matrix
ERR (np.array (same size)) – relative error matrix
IP (np.array(same size)) – induced polarization data matrix
IPERR (np.array(same size)) – induced polarization error matrix
- pygimli.physics.ert.coverageERT()#
coverageDCtrans( (object)S, (object)dd, (object)mm) -> object :
- C++ signature :
GIMLI::Vector<double> coverageDCtrans(GIMLI::MatrixBase,GIMLI::Vector<double>,GIMLI::Vector<double>)
- pygimli.physics.ert.createData(elecs, schemeName='none', **kwargs)[source]#
Utility one-liner to create a BERT datafile
- Parameters:
elecs (int | list[pos] | array(x)) – Number of electrodes or electrode positions or x-positions
schemeName (str ['none']) – Name of the configuration. If you provide an unknown scheme name, all known schemes [‘wa’, ‘wb’, ‘pp’, ‘pd’, ‘dd’, ‘slm’, ‘hw’, ‘gr’] listed.
**kwargs –
Arguments that will be forwarded to the scheme generator.
- inversebool
interchange AB MN with MN AB
- reciprocitybool
interchange AB MN with BA NM
- addInversebool
add additional inverse measurements
- spacingfloat [1]
electrode spacing in meters
- closedbool
Close the chain. Measure from the end of the array to the first electrode.
- Returns:
data
- Return type:
DataContainerERT
Examples
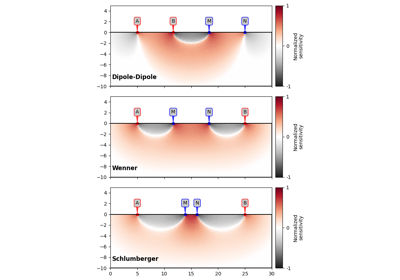
>>> import matplotlib.pyplot as plt >>> from pygimli.physics import ert >>> >>> schemes = ['wa', 'wb', 'pp', 'pd', 'dd', 'slm', 'hw', 'gr'] >>> fig, ax = plt.subplots(3,3) >>> >>> for i, schemeName in enumerate(schemes): ... s = ert.createData(elecs=41, schemeName=schemeName) ... k = ert.createGeometricFactors(s) ... _ = ert.show(s, vals=k, ax=ax.flat[i], label='k - ' + schemeName) >>> >>> plt.show()
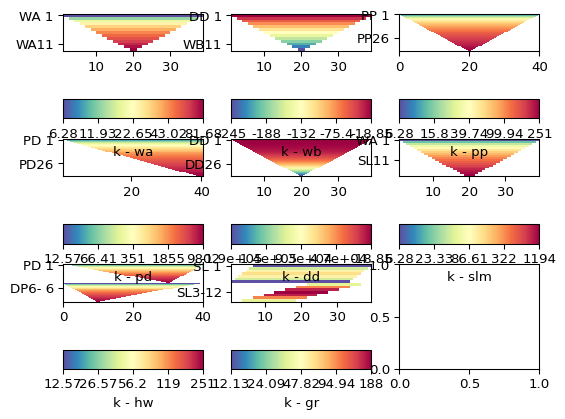
Examples using pygimli.physics.ert.createData
- pygimli.physics.ert.createERTData(*args, **kwargs)#
- pygimli.physics.ert.createGeometricFactors(*args, **kwargs)#
Create geometric factors for a given data scheme.
Create geometric factors for a data scheme with and without topography. Calculation will be done analytical (only for half space geometry) or numerical.
This function caches the result depending on scheme, mesh and pg.version()
- Parameters:
scheme (GIMLI::DataContainerERT) – Datacontainer of the scheme.
numerical (bool | None [False]) – If numerical is None, False is assumed, we try to guess topography and warn if we think we found them. If set to True or False, numerical calculation will used or not.
mesh (GIMLI::Mesh | str) – Mesh for numerical calculation. If not given, analytical geometric factors for halfspace earth are guessed or a default mesh will be created (and h/p refined according to h2/p2). If given topo is set to True. If the numerical effort is to high or the accuracy to low you should consider calculating the factors manually.
h2 (bool [True]) – Default spatial refinement to achieve high accuracy
p2 (bool [True]) – Default polynomial refinement to achieve high accuracy
forceFlatEarth (bool [False]) – Set z values of electrodes to zero for geometric factor (for topo).
verbose (bool) – Give some output.
Examples using pygimli.physics.ert.createGeometricFactors
- pygimli.physics.ert.createInversionMesh(data, **kwargs)[source]#
Create default mesh for ERT inversion.
- Parameters:
data (GIMLI::DataContainerERT) – Data Container needs at least sensors to define the geometry of the mesh.
:param Forwarded to
pygimli.meshtools.createParaMesh:- Returns:
mesh – Inversion mesh with default marker (1 for background, 2 parametric domain)
- Return type:
- pygimli.physics.ert.drawERTData(ax, data, vals=None, **kwargs)[source]#
Plot ERT data as pseudosection matrix (position over separation).
- Parameters:
data (DataContainerERT) – data container with sensorPositions and a/b/m/n fields
vals (iterable of data.size() [data['rhoa']]) – vector containing the vals to show
ax (mpl.axis) – axis to plot, if not given a new figure is created
cMin/cMax (float) – minimum/maximum color vals
logScale (bool) – logarithmic colour scale [min(A)>0]
label (string) – colorbar label
**kwargs –
- dxfloat
x-width of individual rectangles
- indinteger iterable or IVector
indices to limit display
- circularbool
Plot in polar coordinates when plotting via patchValMap
- Returns:
ax – The used Axes
cbar – The used Colorbar or None
- pygimli.physics.ert.estimateError(data, relativeError=0.03, absoluteUError=None, absoluteError=0, absoluteCurrent=0.1)[source]#
Estimate relative error based on relative and absolute parts.
- Parameters:
relativeError (float [0.03]) – relative error level in %/100 for u, R or rhoa
absoluteUError (float [None]) – Absolute potential error in V. Needs ‘u’ values in data. Otherwise calculate them from ‘rhoa’, ‘k’ and absoluteCurrent if no ‘i’ given.
absoluteError (float [0.0]) – Absolute data error in Ohm m. Needs ‘R’ or ‘rhoa’ values.
absoluteCurrent (float [0.1]) – Current level in A for reconstruction for absolute potential V
- Returns:
error
- Return type:
Array
Examples using pygimli.physics.ert.estimateError
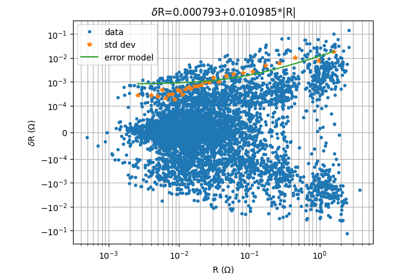
- pygimli.physics.ert.fitReciprocalErrorModel(data, nBins=None, show=False, rel=False)[source]#
Fit an error by statistical normal-reciprocal analysis.
Identify normal reciprocal pairs and fit error model to it
- Parameters:
data (pg.DataContainerERT) – input data
nBins (int) – number of bins to subdivide data (4 < data.size()//30 < 30)
rel (bool [False]) – fit relative instead of absolute errors
show (bool [False]) – generate plot of reciprocals, mean/std bins and error model
- Returns:
ab – indices of a+b*R (rel=False) or a+b/R (rec=True)
- Return type:
[float, float]
Examples using pygimli.physics.ert.fitReciprocalErrorModel
- pygimli.physics.ert.generateDataFromUniqueIndex(ind, data=None, nI=None)[source]#
Generate data container from unique index.
- pygimli.physics.ert.generateDataPDF(data, filename='data.pdf', **kwargs)[source]#
Generate a multi-page pdf showing all data properties.
- pygimli.physics.ert.geometricFactor()#
- geometricFactors( (object)data [, (object)dim=3 [, (object)forceFlatEarth=False]]) -> object :
Helper function to calculate configuration factors for a given DataContainerERT
- C++ signature :
GIMLI::Vector<double> geometricFactors(GIMLI::DataContainerERT [,int=3 [,bool=False]])
- pygimli.physics.ert.geometricFactors((object)data[, (object)dim=3[, (object)forceFlatEarth=False]]) object :#
Helper function to calculate configuration factors for a given DataContainerERT
- C++ signature :
GIMLI::Vector<double> geometricFactors(GIMLI::DataContainerERT [,int=3 [,bool=False]])
- pygimli.physics.ert.load(fileName, verbose=False, **kwargs)[source]#
Shortcut to load ERT data.
Import Data and try to assume the file format. Additionally to unified data format we support the wide-spread res2dinv format as well as ASCII column files generated by the processing software of various instruments (ABEM LS, Syscal Pro, Resecs, ?)
If this fails, install pybert and use its auto importer pybert.importData.
- Parameters:
fileName (str) – file name
verbose (bool) – print some stuff or not
ensureKRhoa (bool) – make sure data container has geometric factors and apparent resistivity
- Returns:
data
- Return type:
pg.DataContainer
- pygimli.physics.ert.reciprocalIndices(data, onlyOnce=False, unify=True)[source]#
Return indices for reciprocal data.
- Parameters:
data (DataContainerERT) – data containing reciprocal data
onlyOnce (bool [False]) – return every pair only once
- Returns:
iN, iR – indices into the data container for normal and reciprocals
- Return type:
np.array(dtype=int)
Examples using pygimli.physics.ert.reciprocalIndices
- pygimli.physics.ert.reciprocalProcessing(data, rel=True, maxrec=0.2, maxerr=0.2)[source]#
Reciprocal data analysis and error estimation.
Carry out workflow for reciprocal * identify normal reciprocal pairs * fit error model to it * estimate error for all measurements * remove one of the pairs (and duplicates) by averaging * filter the data for maximum reciprocity and error
- Parameters:
out (pg.DataContainerERT) – input data container with possible
rel (bool) – base analysis on relative instead of absolute errors
maxrec (float [0.2]) – maximum reciprocity allowed in data
maxerr (float [0.2]) – maximum error estimate
- Returns:
out – filtered data container with pairs removed/averaged
- Return type:
pg.DataContainerERT
Examples using pygimli.physics.ert.reciprocalProcessing
- pygimli.physics.ert.removeDuplicates(data, mode: str = 'average')[source]#
Remove duplicate rows from an ERT datacontainer.
- Parameters:
data (ERTDataContainer) – The data container from which duplicates should be removed.
mode (str, optional) – The method to handle duplicates, choose between - “average” - “first” - “last” - “minerr” - “weighted”
- pygimli.physics.ert.show(data, vals=None, **kwargs)#
Plot ERT data as pseudosection matrix (position over separation).
Creates figure, axis and draw a pseudosection.
- Parameters:
data (DataContainerERT) – Data container with sensorPositions and a/b/m/n fields
vals (array[nData] | str) – Values to be plotted. Default is data[‘rhoa’]. Can be array or string whose data field is extracted.
- Keyword Arguments:
**kwargs –
- axesmatplotlib.axes
Axes to plot into. By default (None), a new figure with a single Axes is created.
- x and ystr | list(str)
forces using matrix plot (drawDataContainerAsMatrix) x, y define the electrode number on x and y axis can be strings (“a”, “m”, “mid”, “sep”) or lists of them [“a”, “m”]
- stylestr
predefined styles for choosing x and y arguments (x/y overrides) - “ab-mn” (default): any combination of current/potential electrodes - “a-m” : only a and m electrode (for unique dipole spacings as DD) - “a-mn” : a and combination of mn electrode (PD with different MN) - “ab-m” : a and combination of mn electrode - “sepa-m” : current dipole length with a and m (multi-gradient) - “a-sepm” : a and potential dipole length with m
- switchxybool
exchange x and y axes before plotting
- Returns:
ax (matplotlib.axes) – axis containing the plots
cb (matplotlib.colorbar) – colorbar instance
Examples using pygimli.physics.ert.show
- pygimli.physics.ert.showData(data, vals=None, **kwargs)#
Plot ERT data as pseudosection matrix (position over separation).
Creates figure, axis and draw a pseudosection.
- Parameters:
data (DataContainerERT) – Data container with sensorPositions and a/b/m/n fields
vals (array[nData] | str) – Values to be plotted. Default is data[‘rhoa’]. Can be array or string whose data field is extracted.
- Keyword Arguments:
**kwargs –
- axesmatplotlib.axes
Axes to plot into. By default (None), a new figure with a single Axes is created.
- x and ystr | list(str)
forces using matrix plot (drawDataContainerAsMatrix) x, y define the electrode number on x and y axis can be strings (“a”, “m”, “mid”, “sep”) or lists of them [“a”, “m”]
- stylestr
predefined styles for choosing x and y arguments (x/y overrides) - “ab-mn” (default): any combination of current/potential electrodes - “a-m” : only a and m electrode (for unique dipole spacings as DD) - “a-mn” : a and combination of mn electrode (PD with different MN) - “ab-m” : a and combination of mn electrode - “sepa-m” : current dipole length with a and m (multi-gradient) - “a-sepm” : a and potential dipole length with m
- switchxybool
exchange x and y axes before plotting
- Returns:
ax (matplotlib.axes) – axis containing the plots
cb (matplotlib.colorbar) – colorbar instance
Examples using pygimli.physics.ert.showData
- pygimli.physics.ert.showERTData(data, vals=None, **kwargs)[source]#
Plot ERT data as pseudosection matrix (position over separation).
Creates figure, axis and draw a pseudosection.
- Parameters:
data (DataContainerERT) – Data container with sensorPositions and a/b/m/n fields
vals (array[nData] | str) – Values to be plotted. Default is data[‘rhoa’]. Can be array or string whose data field is extracted.
- Keyword Arguments:
**kwargs –
- axesmatplotlib.axes
Axes to plot into. By default (None), a new figure with a single Axes is created.
- x and ystr | list(str)
forces using matrix plot (drawDataContainerAsMatrix) x, y define the electrode number on x and y axis can be strings (“a”, “m”, “mid”, “sep”) or lists of them [“a”, “m”]
- stylestr
predefined styles for choosing x and y arguments (x/y overrides) - “ab-mn” (default): any combination of current/potential electrodes - “a-m” : only a and m electrode (for unique dipole spacings as DD) - “a-mn” : a and combination of mn electrode (PD with different MN) - “ab-m” : a and combination of mn electrode - “sepa-m” : current dipole length with a and m (multi-gradient) - “a-sepm” : a and potential dipole length with m
- switchxybool
exchange x and y axes before plotting
- Returns:
ax (matplotlib.axes) – axis containing the plots
cb (matplotlib.colorbar) – colorbar instance
Examples using pygimli.physics.ert.showERTData
- pygimli.physics.ert.simulate(mesh, scheme, res, **kwargs)[source]#
Simulate an ERT measurement.
Perform the forward task for a given mesh, resistivity distribution & measuring scheme and return data (apparent resistivity) or potentials.
For complex resistivity, the data contains an apparent phase or the returned potentials are complex.
The forward operator itself only calculates potential values for the electrodes in the given data scheme. To calculate apparent resistivities, geometric factors (k) are needed. If there are no values k in the DataContainerERT scheme, the function tries to calculate them, either analytically or numerically by using a p2-refined version of the given mesh.
- Parameters:
mesh (GIMLI::Mesh) – 2D or 3D Mesh to calculate for.
res (float, array(mesh.cellCount()), array(N, mesh.cellCount()), list) – Resistivity distribution for the given mesh cells can be: - float for homogeneous resistivity (e.g. 1.0) - single array of length mesh.cellCount() - matrix of N resistivity distributions of length mesh.cellCount() - resistivity map as [[regionMarker0, res0], [regionMarker0, res1],…]
scheme (GIMLI::DataContainerERT) – Data measurement scheme.
- Keyword Arguments:
verbose (bool[False]) – Be verbose. Will override class settings.
calcOnly (bool [False]) – Use fop.calculate instead of fop.response. Useful if you want to force the calculation of impedances for homogeneous models. No noise handling. Solution is put as token ‘u’ in the returned DataContainerERT.
noiseLevel (float [0.0]) – add normally distributed noise based on scheme[‘err’] or on noiseLevel if error>0 is not contained
noiseAbs (float [0.0]) – Absolute voltage error in V
returnArray (bool [False]) – Returns an array of apparent resistivities instead of a DataContainerERT
returnFields (bool [False]) – Returns a matrix of all potential values (per mesh nodes) for each injection electrodes.
sr (bool) – use secondary field (singularity removal)
seed (int) – numpy.random seed for repeatable noise in synthetic experiments
phiErr (float|iterable) – absolute phase error, if not given, data[‘iperr’] or noiseLevel is used
float|iterables (contactImpedances) – contact impedances for being used with CEM model
current (float) – current to be assumed
- Returns:
DataContainerERT | array(data.size()) | array(N, data.size()) |
array(N, mesh.nodeCount()) – Data container with resulting apparent resistivity data[‘rhoa’] and errors (if noiseLevel or noiseAbs is set). Optionally return a Matrix of rhoa values (for returnArray==True forces noiseLevel=0). In case of complex-valued resistivity, phase values are contained in data[‘phia’] or returned as additionally returned array.
Examples
>>> from pygimli.physics import ert >>> import pygimli as pg >>> import pygimli.meshtools as mt >>> world = mt.createWorld(start=[-50, 0], end=[50, -50], ... layers=[-1, -5], worldMarker=True) >>> scheme = ert.createData( ... elecs=pg.utils.grange(start=-10, end=10, n=21), ... schemeName='dd') >>> for pos in scheme.sensorPositions(): ... _= world.createNode(pos) ... _= world.createNode(pos + [0.0, -0.1]) >>> mesh = mt.createMesh(world, quality=34) >>> rhomap = [ ... [1, 100. + 0j], ... [2, 50. + 0j], ... [3, 10.+ 1j], ... ] >>> data = ert.simulate(mesh, res=rhomap, scheme=scheme, verbose=True)
Examples using pygimli.physics.ert.simulate
- pygimli.physics.ert.uniqueERTIndex(data, nI=0, reverse=False, unify=True)[source]#
Generate unique index from sensor indices A/B/M/N for matching.
- Parameters:
data (DataContainerERT) – data container holding a b m n field registered as indices (int)
nI (int [0]) – index to generate (multiply), by default (0) sensorCount if two data files with different sensorCount are compared make sure to use the same nI for both
reverse (bool [False]) – exchange current (A, B) with potential (M, N) for reciprocal analysis
unify (bool [True]) – sort A/B and M/N so that bipole orientation does not matter
Classes#
- class pygimli.physics.ert.CrossholeERT(filename=None, **kwargs)[source]#
Bases:
TimelapseERTClass for crosshole ERT data manipulation.
Note that this class is to be split into a hierarchy of classes for general timelapse data management, timelapse ERT and crosshole ERT. You can load data, filter them data in the temporal or measuring axis, plot data, run inversion and export data and result files.
- __init__(filename=None, **kwargs)[source]#
Initialize class and possibly load data.
- Parameters:
filename (str) – filename to load data, times, RHOA and ERR from
data (DataContainerERT) – The data with quadrupoles for all
times (np.array of datetime objects) – measuring times
DATA (2d np.array (data.size(), len(times))) – all apparent resistivities
ERR (2d np.array (data.size(), len(times))) – all apparent relative errors
bhmap (array) – map electrode numbers to borehole numbers
mesh (array) – mesh for inversion
- createMesh(ibound=2, obound=10, quality=None, show=False, threeD=None, ref=0.25, area=1)[source]#
Create a 2D mesh around boreholes.
- Parameters:
ibound (float) – inner boundary in m
ibound – outer boundary in m
quality (float) – triangle or tetgen quality
threeD (bool|None) – create 3D model (None-automatic)
ref (float) – electrode refinement in m
- extractSubset(nbh, plane=None, name=None)[source]#
Extract a subset (slice) by borehole number.
Returns a CrossholeERT instance with reduced boreholes
- Parameters:
nbh (int|array) – borehole(s) to extract data from
name (str) – name to give the new instance
plane (bool [None]) – reduce to 2D (automatic if max. 2 boreholes are used)
- showData(v='rhoa', x='a', y='m', t=None, **kwargs)[source]#
Show data.
Show data as pseudosections (single-hole) or cross-plot (crosshole)
- Parameters:
v (str|array ["rhoa]) – array or field to plot
x (str|array ["a", "m"]) – values to use for x and y axes
y (str|array ["a", "m"]) – values to use for x and y axes
t (int|str|datetime) – time to choose
kwargs (dict) – forwarded to ert.show or showDataContainerAsMatrix
- pygimli.physics.ert.DataContainer#
alias of
DataContainerERT
- class pygimli.physics.ert.ERTIPManager(*args, **kwargs)[source]#
Bases:
ERTManagerMethod manager for ERT including induced polarization (IP).
This class should be use for any single IP data, which can be a single-frequency frequency-domain (FD) amplitude and phase, or a time-domain (TD) IP chargeability (one gate or an integral value).
- __init__(*args, **kwargs)[source]#
Initialize DC part of it (parent class).
- Parameters:
fd (bool) – Frequency-domain, otherwise time-domain
- showFit(**kwargs)[source]#
Show data fit for both app. res and IP.
Forwards to showDCFit and showIPFit
- Parameters:
ipkw (dict) – dictionary forwarded to showIPFit
Keywords
--------
showDCFit (dictionary forwarded to)
- class pygimli.physics.ert.ERTManager(data=None, **kwargs)[source]#
Bases:
MeshMethodManagerERT Manager.
Method Manager for Electrical Resistivity Tomography (ERT)
- __init__(data=None, **kwargs)[source]#
Create ERT Manager instance.
- Parameters:
data (GIMLI::DataContainerERT | str) – You can initialize the Manager with data or give them a dataset when calling the inversion.
useBert (*) – Use Bert forward operator instead of the reference implementation.
sr (*) – Calculate with singularity removal technique. Recommended but needs the primary potential. For flat earth cases the primary potential will be calculated analytical. For domains with topography the primary potential will be calculated numerical using a p2 refined mesh or you provide primary potentials with setPrimPot.
- checkData(data=None)[source]#
Return data from container.
THINKABOUT: Data will be changed, or should the manager keep a copy?
- checkErrors(err, dataVals)[source]#
Check (estimate) and return relative error.
By default we assume ‘err’ are relative values.
- createMesh(data=None, **kwargs)[source]#
Create default inversion mesh.
Forwarded to
pygimli.physics.ert.createInversionMesh
- estimateError(data=None, **kwargs)[source]#
Estimate error composed of an absolute and a relative part.
- Parameters:
absoluteError (float [0.001]) – Absolute data error in Ohm m. Need ‘rhoa’ values in data.
relativeError (float [0.03]) – relative error level in %/100
absoluteUError (float [0.001]) – Absolute potential error in V. Need ‘u’ values in data. Or calculate them from ‘rhoa’, ‘k’ and absoluteCurrent if no ‘i’ is given
absoluteCurrent (float [0.1]) – Current level in A for reconstruction for absolute potential V
- Returns:
error
- Return type:
Array
- load(fileName)[source]#
Load ERT data.
Forwarded to
pygimli.physics.ert.load- Parameters:
fileName (str) – Filename for the data.
- Returns:
data
- Return type:
- saveResult(folder=None, size=(16, 10), **kwargs)[source]#
Save all results in the specified folder.
- Saved items are:
Inverted profile Resistivity vector Coverage vector Standardized coverage vector Mesh (bms and vtk with results)
- showModel(model=None, ax=None, elecs=True, **kwargs)[source]#
Show the last inversion result.
- Parameters:
model (iterable [None]) – Model vector to be drawn. Default is self.model from the last run
ax (mpl axes) – Axes object to draw into. Create a new if its not given.
elecs (bool) – Draw electrodes
**kwargs (dict) – arguments passed to pg.show (cMin, cMax, cMap, logScale)
- Return type:
ax, cbar
- simulate(*args, **kwargs)[source]#
Simulate an ERT measurement.
Perform the forward task for a given mesh, resistivity distribution & measuring scheme and return data (apparent resistivity) or potentials.
For complex resistivity, the apparent resistivities is complex as well.
The forward operator itself only calculates potential values for the electrodes in the given data scheme. To calculate apparent resistivities, geometric factors (k) are needed. If there are no values k in the DataContainerERT scheme, the function tries to calculate them, either analytically or numerically by using a p2-refined version of the given mesh.
- Parameters:
mesh (GIMLI::Mesh) – 2D or 3D Mesh to calculate for.
res (float, array(mesh.cellCount()) | array(N, mesh.cellCount()) |) – list Resistivity distribution for the given mesh cells can be: - float for homogeneous resistivity (e.g. 1.0) - single array of length mesh.cellCount() - matrix of N resistivity distributions of length mesh.cellCount() - resistivity map [[regionMarker0, res0], [regionMarker0, res1],…]
scheme (GIMLI::DataContainerERT) – Data measurement scheme.
- Keyword Arguments:
verbose (bool[False]) – Be verbose. Will override class settings.
calcOnly (bool [False]) – Use fop.calculate instead of fop.response. Useful if you want to force the calculation of impedances for homogeneous models. No noise handling. Solution is put as token ‘u’ in the returned DataContainerERT.
noiseLevel (float [0.0]) – add normally distributed noise based on scheme[‘err’] or on noiseLevel if error>0 is not contained
noiseAbs (float [0.0]) – Absolute voltage error in V
returnArray (bool [False]) – Returns an array of apparent resistivities instead of a DataContainerERT
returnFields (bool [False]) – Returns a matrix of all potential values (per mesh nodes) for each injection electrodes.
- Returns:
DataContainerERT | array(data.size()) | array(N, data.size()) |
array(N, mesh.nodeCount()) – Data container with resulting apparent resistivity data and errors (if noiseLevel or noiseAbs is set). Optional returns a Matrix of rhoa values (for returnArray==True forces noiseLevel=0). In case of a complex valued resistivity model, phase values are returned in the DataContainerERT (see example below), or as an additionally returned array.
Examples
# >>> from pygimli.physics import ert # >>> import pygimli as pg # >>> import pygimli.meshtools as mt # >>> world = mt.createWorld(start=[-50, 0], end=[50, -50], # … layers=[-1, -5], worldMarker=True) # >>> scheme = ert.createData( # … elecs=pg.utils.grange(start=-10, end=10, n=21), # … schemeName=’dd’) # >>> for pos in scheme.sensorPositions(): # … _= world.createNode(pos) # … _= world.createNode(pos + [0.0, -0.1]) # >>> mesh = mt.createMesh(world, quality=34) # >>> rhomap = [ # … [1, 100. + 0j], # … [2, 50. + 0j], # … [3, 10.+ 0j], # … ] # >>> data = ert.simulate(mesh, res=rhomap, scheme=scheme, verbose=1) # >>> rhoa = data.get(‘rhoa’).array() # >>> phia = data.get(‘phia’).array()
- class pygimli.physics.ert.ERTModelling(sr=True, verbose=False)[source]#
Bases:
ERTModellingBaseForward operator for Electrical Resistivity Tomography.
Note
Convention for complex resistiviy inversion: We want to use logarithm transformation for the imaginary part of model so we need the startmodel to have positive imaginary parts. The sign is flipped back to physical correct assumption before we call the response function. The Jacobian is calculated with negative imaginary parts and will be a conjugated complex block matrix for further calulations.
- __init__(sr=True, verbose=False)[source]#
Initialize.
- Variables:
fop (pg.frameworks.Modelling)
data (pg.DataContainer)
modelTrans ([pg.trans.TransLog()])
- Keyword Arguments:
**kwargs – fop: Modelling
- property parameterCount#
Return number of parameters.
- class pygimli.physics.ert.ERTModellingReference(**kwargs)[source]#
Bases:
ERTModellingBaseReference implementation for 2.5D Electrical Resistivity Tomography.
- __init__(**kwargs)[source]#
Initialize.
- Variables:
fop (pg.frameworks.Modelling)
data (pg.DataContainer)
modelTrans ([pg.trans.TransLog()])
- Keyword Arguments:
**kwargs – fop: Modelling
- pygimli.physics.ert.Manager#
alias of
ERTManager
- pygimli.physics.ert.Modelling#
alias of
ERTModelling
- class pygimli.physics.ert.TimelapseERT(filename=None, **kwargs)[source]#
Bases:
objectClass for crosshole ERT data manipulation.
Note that this class is to be split into a hierarchy of classes for general timelapse data management, timelapse ERT and crosshole ERT. You can load data, filter them data in the temporal or measuring axis, plot data, run inversion and export data and result files.
- __init__(filename=None, **kwargs)[source]#
Initialize class and possibly load data.
- Parameters:
filename (str) – filename to load data, times, RHOA and ERR from
data (DataContainerERT) – The data with quadrupoles for all
times (np.array of datetime objects) – measuring times
DATA (2d np.array (data.size(), len(times))) – all apparent resistivities
ERR (2d np.array (data.size(), len(times))) – all apparent relative errors
bhmap (array) – map electrode numbers to borehole numbers
mesh (array) – mesh for inversion
- automask(dmax=0.3, nc=5, robustData=True)[source]#
Automatic outlier masking using dist to smoothed curve.
- Parameters:
dmax (float) – maximum relative deviation
nc (int) – number of coefficient pairs
- chooseTime(t=None)[source]#
Return data for a specific time index or timestamp.
- Parameters:
t (int, str, or datetime, optional) – The time index (int), time label (str), or datetime object specifying the desired time slice. If None, defaults to the first time index.
- Returns:
data – A copy of the data container with the ‘rhoa’ (apparent resistivity) and ‘err’ (error) fields updated for the selected time. If error data is missing or invalid, error is estimated.
- Return type:
DataContainerERT
Notes
If t is not an integer, it is converted to a time index using self.timeIndex(t).
Handles masked values in “rhoa” by replacing them with the median of the absolute values.
If error data is missing or invalid, data.estimateError() is called to estimate errors.
- filter(tmin=0, tmax=None, t=None, select=None, kmax=None)[source]#
Filter data set temporally or data-wise.
- Parameters:
tmin (int|str|datetime) – minimum and maximum times to keep
tmax (int|str|datetime) – minimum and maximum times to keep
t (int|str|datetime) – time to remove
select (list[int]) – times to select
kmax (float) – maximum geometric factor to allow
- fitReciprocalErrorModel(**kwargs)[source]#
Fit all data by analysing normal/reciprocal data.
- Parameters:
show (bool) – show temporal behaviour of absolute & relative errors
ert.fitReciprocalErrorModel) (kwargs passed on to) –
- nBinsint
number of bins to subdivide data (4 < data.size()//30 < 30)
- relbool [False]
fit relative instead of absolute errors
- Returns:
p, a – relative (p) and absolute (a) errors for every time step
- Return type:
array
- generateDataPDF(**kwargs)[source]#
Generate a pdf with all data as timesteps in individual pages.
Iterates through times and calls showData into multi-page pdf
- generateModelPDF(**kwargs)[source]#
Generate a multi-page pdf with the model results.
- Parameters:
**kwargs (keyword arguments passed to pg.show())
- generateRatioPDF(**kwargs)[source]#
Generate a multi-page pdf with the model results.
- Parameters:
creep (bool [False]) – Use preceding time step as reference (default is baseline)
cMax (float [1/cMax]) – maximum of color scale
cMax – minimum of color scale
logScale (bool [True]) – logarithmic color scale
cMap (str ['bwr']) – colormap
- generateTimelinePDF(key='a', filename=None, **kwargs)[source]#
Generate multipage PDF with timeline data.
- Parameters:
key (str ['a']) – data key to sort measurements after
filename (str [name+'time'+key+'.pdf']) – output pdf filename
kwargs (dict) – passed to showTimeline
- invert(t=None, reg=None, regTL=None, **kwargs)[source]#
Run inversion for a specific timestep or all subsequently.
- Parameters:
t (int|datetime|str|array) – time index, string or datetime object, or array of any of these
reg (dict) – regularization options (setRegularization) for all inversions
regTL (dict) – regularization options for timesteps inversion only
- Keyword Arguments:
**kwargs (dict) – keyword arguments passed to ERTManager.invert
- mask(rmin=0.1, rmax=1000000.0, emax=None)[source]#
Mask data (i.e. remove them from inversion).
- Parameters:
rmin (float) – minimum and maximum apparent resistivity
rmax (float) – minimum and maximum apparent resistivity
emax (float) – maximum error
- saveData(filename=None, masknan=True)[source]#
Save all data as datacontainer, times, rhoa and error arrays.
- showAllModels(ncols=2, **kwargs)[source]#
Show all models as subplots.
- Parameters:
ncols (int [2]) – number of columns
**kwargs (dict) – keyword arguments passed to pg.show
- showData(v='rhoa', t=None, **kwargs)[source]#
Show data as pseudosections (single-hole) or cross-plot (crosshole).
- Parameters:
v (str|array ["rhoa]) – array or field to plot
t (int|str|datetime) – time to choose (can also be first argument)
x (str|array ["a", "m"]) – values to use for x and y axes
y (str|array ["a", "m"]) – values to use for x and y axes
crossplot (bool [x and y given]) – force AB-MN crossplot
kwargs (dict) – forwarded to ert.show or showDataContainerAsMatrix
- showTimeline(what='rho', ax=None, **kwargs)[source]#
Show data timeline.
- Parameters:
ax (mpl.Axes|None) – matplotlib axes to plot (otherwise new)
what (str ["rhoa"]) – define what to plot
a (int) – tokens to extract data from
b (int) – tokens to extract data from
m (int) – tokens to extract data from
n (int) – tokens to extract data from