Note
Go to the end to download the full example code.
Synthetic TDIP modelling and inversion#
This example demonstrates the ERTIPManager introduced in pyGIMLi v1.4.3 for use with time-domain chargeability.
Whereas the classic ERTManager is for apparent resistivity only (that is inverted into a resistivity distribution), the ERTIPManager extends this by a simple IP component. This can be in the frequency domain (FD), i.e. apparent phase angle, or in the time domain, i.e. a chargeability.
Note that the ERT simulation can already simulate with complex (i.e. FD) resistivity, but not invert for it.
import numpy as np
import pygimli as pg
from pygimli.physics import ert
import pygimli.meshtools as mt
Synthetic model#
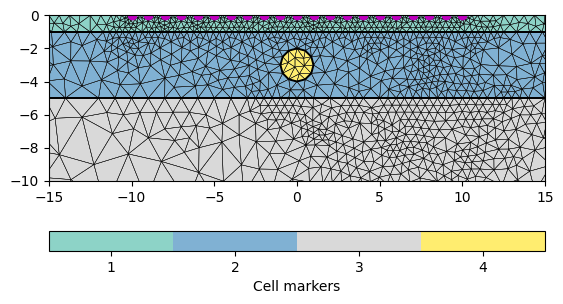
We build a layered model with layer boundaries at 1 and 5m depth. Additionally, we add a circle into the middle layer.
world = mt.createWorld(start=[-50, 0], end=[50, -50],
layers=[-1, -5], worldMarker=True)
scheme = ert.createData(elecs=pg.utils.grange(start=-10, end=10, n=21),
schemeName='dd')
circle = mt.createCircle(pos=[0, -3], radius=1, marker=4)
world += circle
for pos in scheme.sensorPositions():
_= world.createNode(pos)
_= world.createNode(pos + [0.0, -0.1])
mesh = mt.createMesh(world, quality=34.4)
ax, cb = pg.show(mesh, markers=True, showMesh=True, boundaryMarkers=False)
ax.plot(pg.x(scheme), pg.y(scheme), "mo")
ax.set_ylim(-10, 0)
_ = ax.set_xlim(-15, 15)
FD simulation#
We associate different resistivities for the three layers and the identical resistivity for the circle, which is the only body with an imaginary component. First we create an FD data set for comparison using the normal simulate.
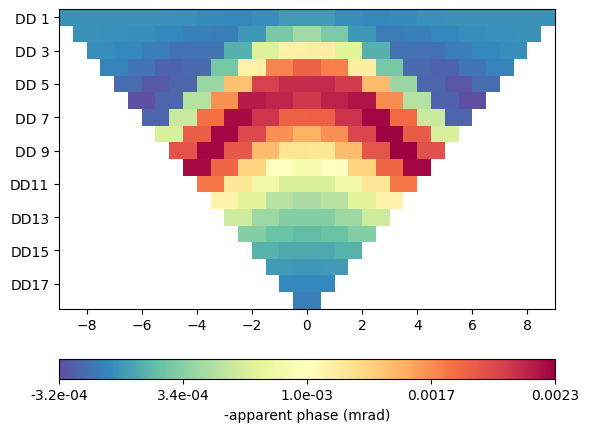
TD simulation#
For time domain, we need a resistivity and a chargeability. Different from before, we specify them as vectors going from 0 to 4 (maximum cell marker). The simulate function first runs a DC forward calculation and then another (AC) computation. The results are stored in the rhoa and ma/ip data fields.
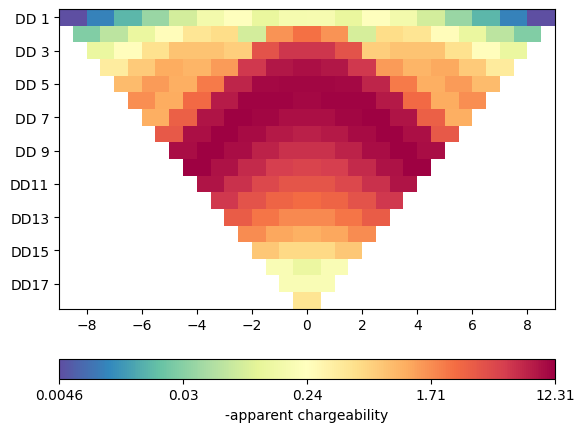
Mesh: Nodes: 1893 Cells: 3521 Boundaries: 5413 3521 3521
Inversion#
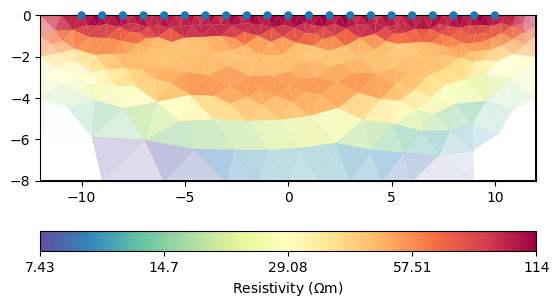
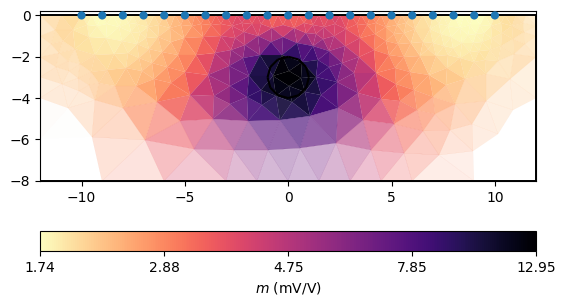
We set a constant error and run an inversion with some keyword arguments. A TDIP inversion can be run by invertTDIP(). showResults show the resistivity result, whereas showIPModel shows the chargeability model. showResults shows both images below each other.
dataTD["err"] = 0.03
mgr = ert.ERTIPManager(dataTD)
mgr.invert(zWeight=0.2, quality=34.4, verbose=True)
mgr.showResult()
ax, cb = mgr.showIPModel()
_ = pg.viewer.mpl.drawPLC(ax, circle, fitView=False,
fillRegion=False, color="white")
--------------------------------------------------------------------------------
--------------------------------------------------------------------------------
inv.iter 1 ... --------------------------------------------------------------------------------
inv.iter 2 ...
################################################################################
# Abort criterion reached: chi² <= 1 (0.69) #
################################################################################
--------------------------------------------------------------------------------
--------------------------------------------------------------------------------
inv.iter 1 ... --------------------------------------------------------------------------------
inv.iter 2 ... --------------------------------------------------------------------------------
inv.iter 3 ... ################################################################################
# Abort criterion reached: dPhi = 1.61 (< 2.0%) #
################################################################################
Total running time of the script: (0 minutes 12.969 seconds)