Note
Go to the end to download the full example code.
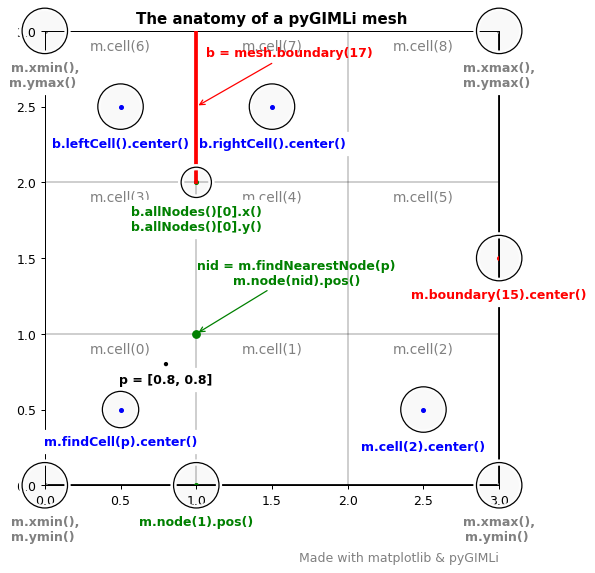
The anatomy of a pyGIMLi mesh#
In this tutorial we look at the anatomy of a GIMLI::Mesh. Although
the example is simplisitic and two-dimensional, the individual members of the
Mesh class and their methods can be inspected similarly for more complex meshes
in 2D and 3D. This example is heavily inspired by the anatomy of a matplotlib
plot, which we can
also highly recommend for visualization with the pygimli.viewer.mpl.
We start by importing matplotlib and defining some helper functions for plotting.
import matplotlib.pyplot as plt
from matplotlib.patches import Circle
from matplotlib.patheffects import withStroke
def circle(x, y, text=None, radius=0.15, c="blue"):
circle = Circle((x, y), radius, clip_on=False, zorder=10, linewidth=1,
edgecolor='black', facecolor=(0, 0, 0, .0125),
path_effects=[withStroke(linewidth=5, foreground='w')])
ax.add_artist(circle)
ax.plot(x, y, color=c, marker=".")
ax.text(x, y - (radius + 0.05), text, backgroundcolor="white",
ha='center', va='top', weight='bold', color=c)
We now import pygimli and create a simple grid/mesh with 3x3 cells.
import pygimli as pg
m = pg.createGrid(4,4)
The following code creates the main plot and shows how the different mesh entities can be called.
# Create matplotlib figure and set size
fig, ax = plt.subplots(figsize=(8,8), dpi=90)
ax.set_title("The anatomy of a pyGIMLi mesh", fontweight="bold")
# Visualize mesh with the generic pg.show command
pg.show(m, ax=ax)
# Number all cells
for cell in m.cells():
node = cell.allNodes()[2]
ax.text(node.x() - 0.5, node.y() - .05, "m.cell(%d)" % cell.id(),
fontsize=11, ha="center", va="top", color="grey")
# Mark horizontal and vertical extent
circle(m.xmin(), m.xmax(), "m.xmin(),\nm.ymax() ", c="grey")
circle(m.xmin(), m.ymin(), "m.xmin(),\nm.ymin() ", c="grey")
circle(m.xmax(), m.ymin(), "m.xmax(),\nm.ymin() ", c="grey")
circle(m.xmax(), m.ymax(), "m.xmax(),\nm.ymax() ", c="grey")
# Mark center of a cell
cid = 2
circle(m.cell(cid).center().x(), m.cell(cid).center().y(),
"m.cell(%d).center()" % cid)
# Mark node
circle(m.node(1).x(), m.node(1).y(), "m.node(1).pos()", c="green")
# Find cell in which point p lies
p = [0.8, 0.8]
circle(p[0], p[1], "p = %s" % p, radius=0.01, c="black")
cell = m.findCell(p)
circle(cell.center().x(), cell.center().y(), "m.findCell(p).center()",
radius=0.12)
# Find closest node to point p
nid = m.findNearestNode(p)
n = m.node(nid)
ax.plot(n.x(), n.y(), "go")
ax.annotate('nid = m.findNearestNode(p)\nm.node(nid).pos()', xy=(n.x(), n.y()),
xycoords='data', xytext=(80, 40), textcoords='offset points',
ha="center", weight='bold', color="green",
arrowprops=dict(arrowstyle='->',
connectionstyle="arc",
color="green"))
# Mark boundary center
bid = 15
boundary_center = m.boundary(bid).center()
circle(boundary_center.x(), boundary_center.y(),
"m.boundary(%d).center()" % bid, c="red")
# Mark boundary together with left and right cell
bid = 17
b = m.boundaries()[bid] # equivalent to mesh.boundary(17)
n1 = b.allNodes()[0]
n2 = b.allNodes()[1]
ax.plot([n1.x(), n2.x()], [n1.y(), n2.y()], "r-", lw=3, zorder=10)
ax.annotate('b = mesh.boundary(%d)' % bid, xy=(b.center().x(), b.center().y()),
xycoords='data', xytext=(8, 40), textcoords='offset points',
weight='bold', color="red",
arrowprops=dict(arrowstyle='->',
connectionstyle="arc",
color="red"))
circle(n1.x(), n1.y(), "b.allNodes()[0].x()\nb.allNodes()[0].y()",
radius=0.1, c="green")
# Mark neighboring cells
left = b.leftCell()
right = b.rightCell()
circle(left.center().x(), left.center().y(),
"b.leftCell().center()", c="blue")
circle(right.center().x(), right.center().y(),
"b.rightCell().center()", c="blue")
ax.text(3.0, -0.5, "Made with matplotlib & pyGIMLi",
fontsize=10, ha="right", color='.5')
fig.tight_layout(pad=5.7)
pg.wait()