Note
Go to the end to download the full example code.
Mesh interpolation#
In this tutorial, we look at the mesh interpolation options in GIMLi. Although the example shown here is in 2D, the same routines can be applied when converting 3D data to a 2D mesh for instance.
Create coarse and fine mesh with data#
def create_mesh_and_data(n):
nc = np.linspace(-2.0, 0.0, n)
mesh = pg.meshtools.createMesh2D(nc, nc)
mcx = pg.x(mesh.cellCenter())
mcy = pg.y(mesh.cellCenter())
data = np.cos(1.5 * mcx) * np.sin(1.5 * mcy)
return mesh, data
coarse, coarse_data = create_mesh_and_data(5)
fine, fine_data = create_mesh_and_data(20)
Interpolate data to different meshes#
We define two functions taking the input mesh, the input data and the output mesh as parameters and return the input data interpolated to the output mesh.
def nearest_neighbor_interpolation(inmesh, indata, outmesh, nan=99.9):
""" Nearest neighbor interpolation. """
outdata = []
for pos in outmesh.cellCenters():
cell = inmesh.findCell(pos)
if cell:
outdata.append(indata[cell.id()])
else:
outdata.append(nan)
return outdata
def linear_interpolation(inmesh, indata, outmesh):
""" Linear interpolation using `pg.interpolate()` """
outdata = pg.Vector() # empty
pg.interpolate(srcMesh=inmesh, inVec=indata,
destPos=outmesh.cellCenters(), outVec=outdata)
# alternatively you can use the interpolation matrix
outdata = inmesh.interpolationMatrix(outmesh.cellCenters()) * \
pg.core.cellDataToPointData(inmesh, indata)
return outdata
Visualization#
meshes = [coarse, fine]
datasets = [coarse_data, fine_data]
ints = [nearest_neighbor_interpolation,
linear_interpolation]
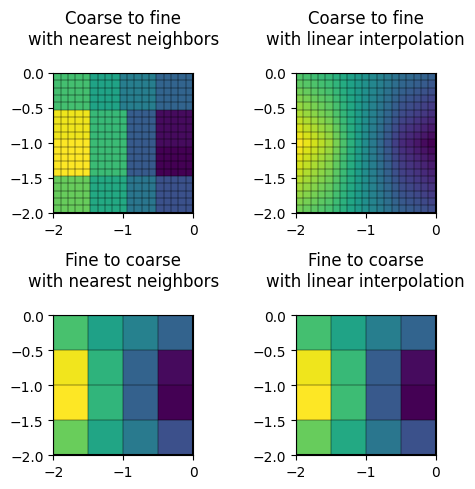
fig, ax = plt.subplots(2, 2, figsize=(5, 5))
# Coarse data to fine mesh
drawModel(ax[0, 0], fine, ints[0](coarse, coarse_data, fine), showCbar=False)
drawMesh(ax[0, 0], fine)
drawModel(ax[0, 1], fine, ints[1](coarse, coarse_data, fine), showCbar=False)
drawMesh(ax[0, 1], fine)
# Fine data to coarse mesh
drawModel(ax[1, 0], coarse, ints[0](fine, fine_data, coarse), showCbar=False)
drawMesh(ax[1, 0], coarse)
drawModel(ax[1, 1], coarse, ints[1](fine, fine_data, coarse), showCbar=False)
drawMesh(ax[1, 1], coarse)
titles = ["Coarse to fine\nwith nearest neighbors",
"Coarse to fine\nwith linear interpolation",
"Fine to coarse\nwith nearest neighbors",
"Fine to coarse\nwith linear interpolation"]
for a, title in zip(ax.flat, titles):
a.set_title(title + "\n")
fig.tight_layout()
plt.show()