2. Fundamentals#
2.1. Software design#
In applied geophysics, a lot of research efforts are directed towards the integration of different physical models and combined inversion approaches to estimate multi-physical subsurface parameters. Such approaches often require coupling of different software packages and file-based data exchange. The idea of pyGIMLi is to present a very flexible framework for geophysical modelling and inversion, which allows standard and customized modelling and inversion workflows to be realized in a reproducible manner.
The software is written in Python on top of a C++ core library, which allows a combination of flexible scripting and numerical efficiency. pyGIMLi uses selected external dependencies for quality constrained mesh generation in 2D Triangle and 3D Tetgen and visualization in 2D (Matplotlib) and 3D (Paraview) for example. For solving linear systems we use the open-source collection SuiteSparse, which contains multi-frontal direct and iterative solvers as well as reordering algorithms.

pyGIMLi is organized in three different abstraction levels:
In the application level, ready-to-use method managers and frameworks are provided. Method managers (pygimli.manager) hold all relevant functionality related to a geophysical method. A method manager can be initialized with a data set and used to analyze and visualize this data set, create a corresponding mesh, and carry out an inversion. Various method managers are available in pygimli.physics. Frameworks (pygimli.frameworks) are generalized abstractions of standard and advanced inversions tasks such as time-lapse or joint inversion for example. Since frameworks communicate through a unified interface, they are method independent.
In the modelling level, users can set up customized forward operators that map discretized parameter distributions to a data vector. Once defined, it is straightforward to set up a corresponding inversion workflow or combine the forward operator with existing ones.
The underlying equation level allows to directly access the finite element (pygimli.solver.solveFiniteElements()) and finite volume (pygimli.solver.solveFiniteVolume()) solvers to solve various partial differential equations on unstructured meshes, i.e. to approach various physical problems with possibly complex 2D and 3D geometries.
2.2. Module/method overview#
Frequency-domain (FD) or time-domain (TD) semi-analytical 1D solutions |
|
Electrical Resistivity Tomography (ERT) |
|
Solve gravimetric and magneto static problems in 2D and 3D analytically |
|
Various petrophysical models |
|
Full wave form seismics utilities and simulations |
|
Spectral induced polarization (SIP) measurements and fittings. |
|
Surface nuclear magnetic resonance (NMR) data inversion |
|
Refraction seismics or first arrival traveltime calculations. |
|
Direct current electromagnetics |
method |
Forward |
Inverse |
Dimension |
|---|---|---|---|
✓ |
✓ |
1D |
|
✓ |
✓ |
2D, 3D, 4D |
|
✓ |
✓ |
2D, 3D |
|
✓ |
✓ |
2D, 3D |
|
✓ |
✓ |
dimensionless |
|
✓ |
- |
“1D” |
|
✓ |
✓ |
1D, 2D |
|
✓ |
✓ |
1D |
|
✓ |
✓ |
2D, (3D) |
|
✓ |
✓ |
1D |
Basic pyGIMLi classes
|
alias of |
|
alias of |
|
alias of |
|
alias of |
|
Note
Please add docstrings for abovementioned classes and then use autosummary function for user guide!
pyGIMLi class |
Description |
|---|---|
|
All elements are stored column-wise, i.e. all rows A[i] are of type |
|
One dimensional array aka Vector of limited size to store data, like ERT sensitivities. |
|
Used for numerical approximation of partial differential equations like finite-element or finite volume. Not typically used unless efficiency is of importance. It exists also complex-valued as pg.matrix.CSparseMatrix |
|
Arbitrary matrices are combined into a logical matrix. This is of importance for inversion frameworks, e.g., concatenated Jacobian matrices during joint inversions. |
|
Data container storing the individual data values as well as any description how they were obtained, e.g. the geometry of source and receivers. |
2.3. Viewer interface#
In pygimli we provide some basic post-processing routines using the matplotlib visualization framework. The main visualization call is pygimli.viewer.show() which is sufficient for most meshes, fields, models and streamline views. It forwards the object to be plotted to a known visualization function. pygimli.viewer.showMesh() is the typical call for the visualization of 2D data.
However, the underlying show functions only provide an input instance and are not directly responsible for plotting the data. Depending on the data type, pg.viewer.showMesh() then again forwards to the following instances of pg.viewer.mpl:
Data type |
Draw function |
|---|---|
2D meshes |
|
Value per mesh cell - model patch |
|
Value per mesh node - scalar field |
|
Iterable of type |
|
|
|
|
An empty show call creates an empty ax window:
Show code cell source
import pygimli as pg
import pygimli.meshtools as mt
world = mt.createWorld(start=[-10, 0], end=[10, -10],
layers=[-3, -7], worldMarker=True)
mesh = mt.createMesh(world, quality=32, area=0.2, smooth=[1, 10])
res = [[1,10],[2,100],[3,999]]
cell_markers = mesh.cellMarkers()
resistivity = [res[marker-1][1] for marker in cell_markers]
nodeDta = pg.meshtools.cellDataToNodeData(mesh, resistivity)
pg.show()
(<AxesSubplot: >, None)
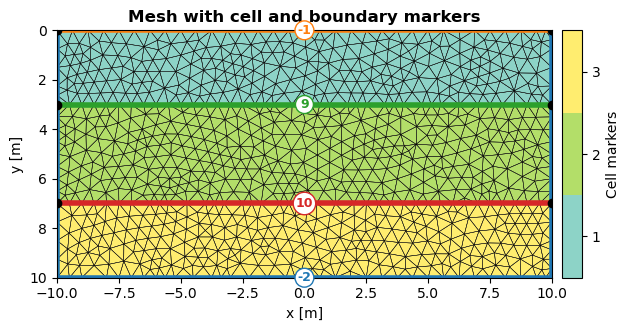
The functions pygimli.viewer.show() and pygimli.viewer.showMesh() return the matplotlib axis object as well as the colorbar object. This allows for further customization of the plot. The following example plots the mesh with cell and boundary markers as “minimal working example”. The orientation of the colorscale can either be parallel to the x- (orientation="horizontal") or y-axis (orientation="vertical"). Further customization of axes or the colorbar can be done by accessing the returned objects directly:
Show code cell source
ax,cbar = pg.viewer.show(mesh, markers=True, showMesh=True, orientation="vertical")
ax.set_title("Mesh with cell and boundary markers", fontweight="bold")
ax.set_xlabel("x [m]")
ax.set_ylabel("y [m]")
Text(46.972222222222186, 0.5, 'y [m]')
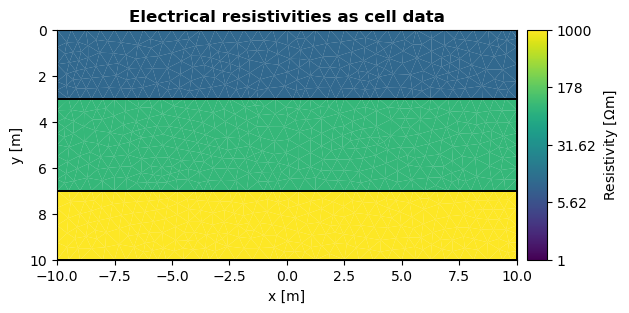
If you want to plot data on top of a mesh, you can use the data argument. The following example shows electrical resistivities as cell data:
Show code cell source
ax, cbar = pg.viewer.show(mesh, data=res, logScale=True, cMin=1, cMax=1000, orientation="vertical", label="Resistivity [$\Omega$m]")
ax.set_title("Electrical resistivities as cell data", fontweight="bold")
ax.set_xlabel("x [m]")
ax.set_ylabel("y [m]")
Text(46.972222222222186, 0.5, 'y [m]')
If you want to plot node data, this can be done with the same argument data. The following example shows the same electrical resistivities as node data:
Show code cell source
ax, cbar = pg.viewer.show(mesh, data=nodeDta, logScale=True, cMin=1, cMax=1000, orientation = "vertical", label="Resistivity [$\Omega$m]")
ax.set_title("Electrical resistivities as node data", fontweight="bold")
ax.set_xlabel("x [m]")
ax.set_ylabel("y [m]")
Text(46.972222222222186, 0.5, 'y [m]')
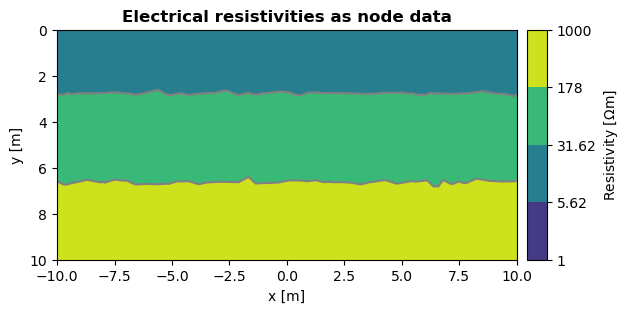
Convert cell to node data
By using the function pg.meshtools.cellDataToNodeData() you can convert cell data to node data. Please refer to the documentation for further details.