Note
Go to the end to download the full example code
Field data inversion (“Koenigsee”)#
This minimalistic example shows how to use the Refraction Manager to invert a field data set. Here, we consider the Koenigsee data set, which represents classical refraction seismics data set with slightly heterogeneous overburden and some high-velocity bedrock. The data file can be found in the pyGIMLi example data repository.
# We import pyGIMLi and the traveltime module.
import matplotlib.pyplot as plt
import pygimli as pg
import pygimli.physics.traveltime as tt
The helper function pg.getExampleData downloads the data set to a temporary location and loads it. Printing the data reveals that there are 714 data points using 63 sensors (shots and geophones) with the data columns s (shot), g (geophone), and t (traveltime). By default, there is also a validity flag.
data = pg.getExampleData("traveltime/koenigsee.sgt", verbose=True)
print(data)
[::::::::::::::::::::::::::::::::::::: 83% ::::::::::::::::::::::: ] 8193 of 9844 complete
[:::::::::::::::::::::::::::::::::::: 100% ::::::::::::::::::::::::::::::::::::] 9844 of 9844 complete
md5: 641890bb17cb2bdf052cbc348669dfd0
Data: Sensors: 63 data: 714, nonzero entries: ['g', 's', 't', 'valid']
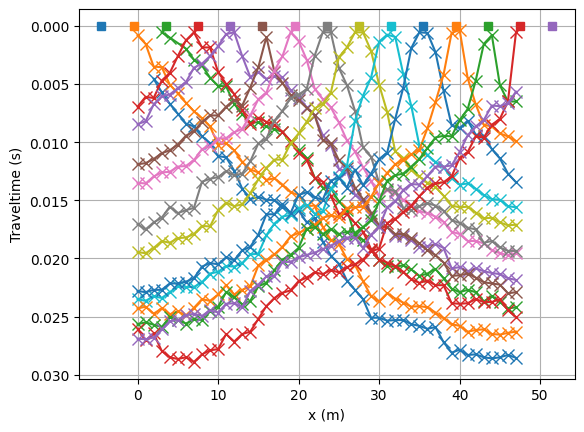
Let’s have a look at the data in the form of traveltime curves.
fig, ax = plt.subplots()
lines = tt.drawFirstPicks(ax, data)
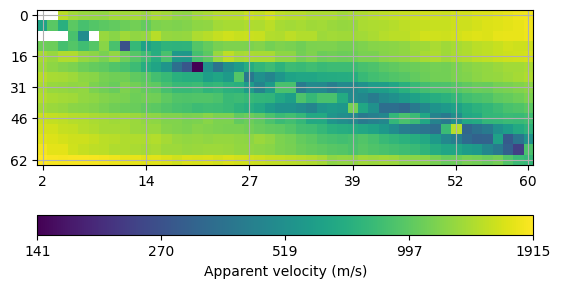
We initialize the refraction manager.
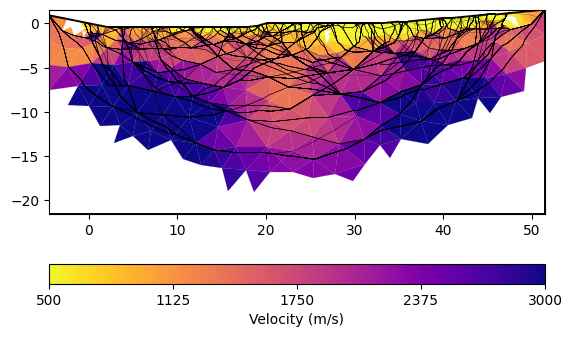
Finally, we call the invert method and plot the result.The mesh is created based on the sensor positions on-the-fly.
mgr.invert(secNodes=3, paraMaxCellSize=5.0,
zWeight=0.2, vTop=500, vBottom=5000, verbose=1)
fop: <pygimli.physics.traveltime.modelling.TravelTimeDijkstraModelling object at 0x7f7bb4355df0>
Data transformation: <pygimli.core._pygimli_.RTrans object at 0x7f7ba1a655d0>
Model transformation (cumulative):
0 <pygimli.core._pygimli_.RTransLogLU object at 0x7f7bb41ac9a0>
min/max (data): 3.5e-04/0.03
min/max (error): 3%/3%
min/max (start model): 2.0e-04/0.002
--------------------------------------------------------------------------------
inv.iter 0 ... chi² = 156.33
--------------------------------------------------------------------------------
inv.iter 1 ... chi² = 12.31 (dPhi = 91.50%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 2 ... chi² = 8.91 (dPhi = 27.38%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 3 ... chi² = 6.58 (dPhi = 25.62%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 4 ... chi² = 5.50 (dPhi = 15.72%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 5 ... chi² = 4.95 (dPhi = 9.28%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 6 ... chi² = 4.45 (dPhi = 9.38%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 7 ... chi² = 4.10 (dPhi = 6.93%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 8 ... chi² = 3.86 (dPhi = 5.29%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 9 ... chi² = 3.74 (dPhi = 2.69%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 10 ... chi² = 3.61 (dPhi = 2.98%) lam: 20.0
--------------------------------------------------------------------------------
inv.iter 11 ... chi² = 3.55 (dPhi = 1.41%) lam: 20.0
################################################################################
# Abort criterion reached: dPhi = 1.41 (< 2.0%) #
################################################################################
1090 [962.1769547915529,...,2676.323601442217]
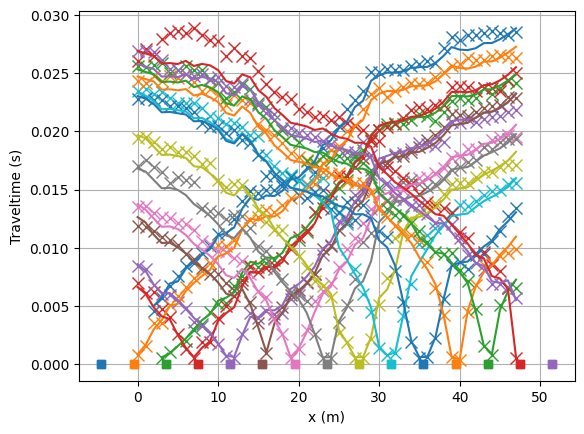
Look at the fit between measured (crosses) and modelled (lines) traveltimes.
mgr.showFit(firstPicks=True)
You can plot only the model and customize with a bunch of keywords
You can play around with the gradient starting model (vTop and vBottom arguments) and the regularization strength lam and customize the mesh.
Total running time of the script: (0 minutes 25.672 seconds)